IBST
Croix du sud 4-5/L7.07.06
1348 Louvain-la-Neuve
Pascal Hols
Professeur
Directeur de recherche FNRS
- Diploma
Year Label Educational Organization 1987 Licencié en Sciences Biologiques Facultés Universitaires Notre Dame de la Paix à Namur 1991 Diplômé d'études approfondies en biologie Université catholique de Louvain 1994 Docteur en sciences Université catholique de Louvain
RESEARCH OVERVIEW
Research activities are focused on a specific group of Gram-positive bacteria, generically referred to as "lactic acid bacteria" (LAB), which are of major industrial importance in food fermentation. Moreover, some LAB species are natural members of the intestinal microflora of mammals where they play a beneficial health role.
Three model species are currently studied at the genetic and physiological levels:
- Lactobacillus plantarum
- Lactococcus lactis
- Streptococcus thermophilus
A multidisciplinary range of genomics/post-genomics, biochemical, and biophysical approaches are used to study the function of genes that are involved in:
Carbon metabolism
Cell wall biosynthesis
Metabolic adaptation to environmental parameters
In order to improve our knowledge of LAB metabolism, we established the complete genome sequence of Streptococcus thermophilus, a yoghurt starter and an unique example of a non-pathogenic Streptococcus.
DNA microarrays are currently exploited for transcriptomic analyses in order to study regulatory networks (quorum-sensing mechanisms for bacteriocin production and competence, nitrogen metabolism, SOS response,...)
Metabolic engineering and heterologous expression technologies are also used to engineer LAB strains to serve as starters in dairy fermentation, or as host systems for the production and the delivery of specific compounds of food and pharmaceutical interest.
RESEARCH ACTIVITIES
Carbon metabolism, genetic control and metabolic engineering
Homofermentative LAB have a relatively simple metabolism completely focused on the rapid conversion of sugars to lactic acid.
Other products are also formed as by-products, such as acetic acid, acetaldehyde, ethanol, and diacetyl, all contributing to the specific flavour of fermented food products. There is almost no overlap between nitrogen and carbon metabolism due to a limited biosynthetic capacity and a limited effect of oxygen on central metabolism (no obvious respiration and Krebs cycle). For all these reasons, these microorganisms are considered as ideal targets of metabolic engineering.
The study of carbon metabolism in homofermentative LAB is mainly focused on pyruvate/glycolytic intermediates dissipating enzymes.
Fundamental studies are focused on the functional role of the following enzymes: lactate dehydrogenases, lactate racemase, lactate oxidases, pyruvate oxidases, NADH oxidases, mannitol dehydrogenases, sorbitol dehydrogenases.
- Gene cloning, inactivation, overexpression
- Transcriptional regulation by oxygen, carbon source, ...
- Physiological role of these enzymes: maintenance of the redox balance, maintenance of the proton gradient through ATP production and survival in stationary phase, interconnection between D-lactate production and cell wall biosynthesis
Concerning applications, we have demonstrated the potential of recombinant strains regarding to pyruvate/glycolytic intermediate rerouting for the production of:
- Lactic acid isomers
- Aroma compounds (acetate, acetaldehyde, diacetyl)
- Low calorie sugars/sweeteners (mannitol, sorbitol, alanine)
We have also assessed the potential of recombinant lactobacilli for the scavenging of intestinal ammonium in several hyperammoniemic mouse models of hepatic encephalopathy.


Key publications:
- BIENERT G.P., DESGUIN B., CHAUMONT F., HOLS P. Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. Biochem. J. 2013, in press.
- TSUJI A., OKADA S., HOLS P., SATOH E. Metabolic engineering of Lactobacillus plantarum for succinic acid production through activation of the reductive branch of the tricarboxylic acid cycle. Enzyme Microb. Technol. 53, 2013, 97-103.
- NICAISE C, PROZZI D, VIAENE E, MORENO C, GUSTOT T, QUERTINMONT E, DEMETTER P, SUAIN V, GOFFIN P, DEVIÈRE J, HOLS P, Control of acute, chronic and constitutive hyperammonemia by wild-type and gentically engineered Lactobacillus plantarum in rodens, Hepatology, 48, 2008, 1184-1192. [PDF]

- LADERO V, RAMOS A, WIERSMA A, GOFFIN P, SCHANCK A, KLEEREBEZEM M, HUGENHOLTZ J, SMID E, HOLS P, High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering, Appl. Environ. Microbiol., 73, 2007, 1864-1872. [PDF]

- GOFFIN P, DEGHORAIN M, MAINARDI J-L, TYTGAT I, CHAMPOMIER-VERGES M-C, KLEEREBEZEM M, HOLS P, Lactate racemization as a rescue pathway for supplying D-lactate to the cell wall biosynthesis machinery in Lactobacillus plantarum, J. Bacteriol., 187, 2005, 6750-6761. [PDF]

- HOLS P, KLEEREBEZEM M, SCHANCK A, FERAIN T, HUGENHOLTZ J, DELCOUR J, DE VOS W, Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering, Nat. Biotechnol., 17, 1999, 588-592. [PDF]

Biosynthesis of the cell wall, control of lysis and interactions with the host

The cell wall of Gram-positive bacteria (including LAB) contains four components (peptidoglycan (PG), teichoic acids (TA), polysaccharides (PS), S-layer [optional] and cell wall associated proteins). The primary role of the cell wall is to resist to turgor pressures of the order of 20 atm in Gram-positive bacteria. A second major function of the cell wall is as an interface between the microorganism and its environment performing numerous interactions mediated by the general biophysical properties of the cell wall (e.g. hydrophobicity, charge) and/or specific cell wall compounds (e.g. PG fragments/TA and immunomodulation, surface proteins and adhesion to epithelial cells).
We have adopted a genetic approach to better understand the functional role of the different constituents of the cell wall in Lactobacillus plantarum and Lactococcus lactis by affecting their biosynthesis through targeted gene inactivation.
Our current researches in this area are:
- Modification in size and composition of peptidoglycan precursors: functional role of alanine racemase, glutamate racemase, lactate racemase, D-Ala-D-Ala dipeptidase, D-Ala-D-Ala[or D-Lac] ligase
- D-alanylation and glycosylation of teichoic acids regarding to the action of autolysins: functional role the dlt operon (D-alanylation) and tagE genes (glucosylation)
- Role of peptidoglycan hydrolases
- Role of post-modifications of peptidoglycan: O-acetylation, amidation
Various phenotypic traits of the resulting mutants are analysed: growth, autolysis, cell morphology, resistance/sensitivity to antibiotics, biophysical properties, chemical composition of cell wall compounds and their precursors, interactions with the host [e.g. adhesion to epithelial cells, in vivo persistence in the GI tract of animal models].
A number of collaborations with experts in the field were developed to set up some of these advanced analyses (Y. Dufrêne [UCL/ISV], M. Kleerebezem [Nl], M.-P. Chapot-Chartier [Fr], J Kok [Nl], J. Errington [UK]).
Concerning applications, some of the constructed mutants were shown to be more immunogenic when used as live delivery vehicles for oral immunisation. Mutants affected in D-alanylation of TA displayed interesting immunomodulatory properties, they are stimulating IL-10 production (anti-inflammatory cytokine) and have a preventive effect in a mice model of inflammatory bowel disease.


Key publications:
- ROLAIN T., BERNARD E., BEAUSSART A., DEGAND H., COURTIN P., EGGE-JACOBSEN W., BRON P.A., MORSOMME P., KLEEREBEZEM M., CHAPOT-CHARTIER M.P., DUFRENE Y.F., HOLS P. O-glycosylation as a novel control mechanism of peptidoglycan hydrolase activity. J. Biol. Chem. 2013, in press.
- ROLAIN T., BERNARD E., COURTIN P., BRON P.A., KLEEREBEZEM M., CHAPOT-CHARTIER M.P., HOLS P. Identification of key peptidoglycan hydrolases for morphogenesis, autolysis, and peptidoglycan composition of Lactobacillus plantarum WCFS1. Microb. Cell Fact. 11, 2012, 137.
- BERNARD E., ROLAIN T., COURTIN P., GUILLOT A., LANGELLA P., HOLS P.*, CHAPOT-CHARTIER M.P.* Characterization of O-acetylation of N-acetylglucosamine: a novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286, 2011, 23950-23958.
- ANDRE G., DEGHORAIN M., BRON P.A., VAN SWAM I.I., KLEEREBEZEM M., HOLS P., DUFRENE Y.F. Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem. Biol. 6, 2011, 366-376.
- ANDRE G., KULAKAUSKAS S., CHAPOT-CHARTIER M.P., NAVET B., DEGHORAIN M., BERNARD E., HOLS P., DUFRENE Y.F. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat .Commun. 1, 2010, 27.
- DELCOUR J, FERAIN T, DEGHORAIN M, PALUMBO E, HOLS P, The biosynthesis and functionality of the cell-wall of lactic acid bacteria, Antonie van Leeuwenhoek, 76, 1999, 156-184. [PDF]

Metabolic adaptation to environmental parameters and gene regulation
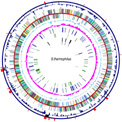
We sequenced the first genome of Streptococcus thermophilus (strain LMG18311, 1.8 Mb, collaboration with Prof. A. Goffeau, Prof. M. Boutry [IS/UCL] and Dr. S. D. Ehrlich [INRA, Fr]) in order to exploit post-genomics tools for the study of transcriptional regulatory networks dealing with the adaptive response to environmental parameters.
DNA microarrays are routinely used and the requested bioinformatics tools for data storage and analysis were implemented.
The genome sequence has revealed a strong adaptation of S. thermophilus to its ecological niche (milk) with a reduced capacity in sugar catabolism and an increased capacity in nitrogen assimilation. Furthermore, most of the genes involved in the pathogenicity of streptococci are absent or inactivated. Its innocuous nature, its exploitation in controlled fermentations and its adaptation to grow in milk suggest the development of a unique specificity in its response to environmental parameters.
Our current researches in this area are:
- Functional role of global regulation circuits: carbon metabolism, nitrogen metabolism, SOS response, ...
- Functional role of two-components regulatory systems and quorum-sensing: bacteriocin production, biofilm formation and competence
A comparative analysis of similar regulatory networks studied by global approaches in other Gram-positive bacteria (Bacillus subtilis, closely related LAB, and pathogenic streptococci) will allow to establish common regulatory pathways and to reveal key target genes in the adaptive response of S. thermophilus. The ultimate goal is to get a comprehensive view at the cell level of the most important parameters controlling metabolism adaptation in this microbe.

Key publications:
- FONTAINE L., GOFFIN P., DUBOUT H., DELPLACE B., BAULARD A., LECAT-GUILLET N., CHAMBELLON E., GARDAN R., HOLS P. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87, 2013, 1113-1132.
- BOUTRY C., WAHL A., DELPLACE B., CLIPPE A., FONTAINE L., HOLS P. Adaptor protein MecA is a negative regulator of the expression of late competence genes in Streptococcus thermophilus. J. Bacteriol. 194, 2012, 1777-1788.
- FONTAINE L., BOUTRY C., DE FRAHAN M.H., DELPLACE B., FREMAUX C., HORVATH P., BOYAVAL P., HOLS P. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 2010, 1444-1454.
- FONTAINE L, BOUTRY C, GUEDON E, GUILLOT A, IBRAHIM M, GROSSIORD B, HOLS P, Quorum-Sensing regulation of the production of Blp bacteriocins in Streptococcus thermophilus, J. Bacteriol., 189, 2007, 7195-7205. [PDF]

- HOLS P, HANCY F, FONTAINE L, GROSSIORD B, PROZZI D, LEBLOND-BOURGET N, DECARIS B, BOLOTIN A, DELORME C, ERHLICH S, GUEDON E, MONNET V, RENAULT P, KLEEREBEZEM M, New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics, FEMS Microbiol. Rev., 29, 2005, 435-463. [PDF]

- BOLOTIN A, QUINQUIS B. , RENAULT P. , SOROKIN A, ERHLICH S, KULAKAUSKAS S, LAPIDUS S, GOLSTMAN E, MAZUR M, PUSCH G, FONSTEIN M, OVERBEEK R, KYPRIDES N, PURNELLE B, PROZZI D, NGUI K, MASUY D, HANCY F, BURTEAU S, BOUTRY M, DELCOUR J, GOFFEAU A, HOLS P, Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus, Nat. Biotechnol., 22, 2004, 1554-1558. [PDF] [PDF]

SCIENTIFIC COLLABORATIONS (RESEARCH ATICLES OR PROJECTS)
 | Carbon metabolism and metabolic engineering in LAB |
| Prof. André N. SCHANCK | UCL | Belgium |
| Prof. Michiel KLEEREBEZEM | ITFN-NIZO food research, Ede | The Netherlands |
| Dr. Jeroen HUGENHOLTZ | Kluyver Centre-NIZO food research, Ede | The Netherlands |
| Prof. Willem M. de VOS | Wageningen Agricultural University | The Netherlands |
| Dr. François LEULIER | Ecole normale supérieure de Lyon | France |
| Prof. Helena SANTOS | ITQB-University of Lisboa, Oieras | Portugal |
| Prof. Margherita SACCO | University of Naples | Italy |
 | Cell wall biosynthesis and degradation in LAB |
| Prof. Yves DUFRÊNE | UCL/ISV | Belgium |
| Drs. Marie-Pierre CHAPOT-CHARTIER, Saulius KULAKAUSKAS, Philippe LANGELLA, and Eric GUEDON | INRA, Jouy en Josas | France |
Profs. R. DANIEL and | University of New Castle upon Tyne | United Kingdom |
| Prof. Oscar KUIPERS | University of Groeningen | The Netherlands |
| Prof. Pier Sandro COCCONCELLI | University of Piacenza | Italy |
| Prof. Thomas HARTUNG | ECVAM-EU Joint Research centre, Ispra | Italy |
| Prof. Armin GEYER | University of Regensburg | Germany |
| Dr. Jean-Luc MAINARDI | INSERM, Paris | France |
| Dr. Sylvie LORTAL | INRA, Rennes | France |
 | LAB as probiotic or live vehicles for the delivery of antigens/enzymes |
| Prof. Jos VANDERLEYDEN | KUL, Leuven | Belgium |
| Prof. Jacques DEVIERE | ULB/Erasme, Bruxelles | Belgium |
| Dr. Annick MERCENIER | Institut Pasteur de Lille | France |
 | Genomics and post-genomics of Streptococcus thermophilus |
Profs. Marc BOUTRY and André GOFFEAU | UCL/ISV | Belgium |
| Dr. Dusko S. EHRLICH | INRA, Jouy en Josas | France |
| Dr. Pierre RENAULT | INRA, Jouy en Josas | France |
| Drs. Véronique MONNET and Rozenn GARDAN | INRA, Jouy en Josas | France |
| Dr. Alain BAULARD | Institut Pasteur Lille | France |
| Prof. Jean-Luc GAILLARD | University of Nancy | France |
| Prof. Bernard DECARIS | University of Nancy | France |
| Dr. Benoît GROSSIORD | ENITA-Bordeaux | France |
EQUIPMENTS
Basic equipment for molecular biology and microbiology (PCR, electrophoresis, gel reader, spectrophotometers, electroporator, incubators, laminar flows,...)
Microarray facilities (Agilent)
DNA microarray scanner and bioinformatics tools for analysis and storage
Nanodrop spectrophotometer for high-fidelity quantitation and purity analysis of DNA and RNA samples
Capillary electrophoresis system (Bioanalyser) for RNA quality control
Small scale fermentors of 2 L and 10 L (control of t°, pH, 02, foam, agitation)
High performance liquid chromatography (HPLC) for the analysis of:
Fermentation products (acetate, lactate, citrate, ethanol, acetoin, butanediol)
Amino-acids (alanine)
Sugars (glucose, fructose, maltose, sucrose, lactose, sorbitol, mannitol)
Lactate isomers (D/L)
Peptidoglycan percursors